Using VMD to draw molecular orbitals
Published:
A quick tutorial on using VMD to visualize molecular orbitals, or other type of orbitals, defined as a vector (linear combination coefficients of the basis set.)
Installation
VMD is free to download from the UIUC Theoretical and Computational Biophysics Group website. I installed Version 1.9.3, as recommended. The installation is standard.
Generating a cube file with PySCF
VMD supports many file types. Here we focus on the .cube files, which is used in Gaussian09. In PySCF, there is a module in tools called cubegen.py, which can produce .cube files storing the electronic density, orbital and molecular electrostatic potential. Here is an example:
from pyscf import scf
from pyscf.tools import cubegen
mol = gto.M(atom='''O 0.00000000, 0.000000, 0.000000
H 0.761561, 0.478993, 0.00000000
H -0.761561, 0.478993, 0.00000000''', basis='6-31g*')
mf = scf.RHF(mol).run()
cubegen.density(mol, 'h2o_den.cube', mf.make_rdm1()) #makes total density
cubegen.mep(mol, 'h2o_pot.cube', mf.make_rdm1())
cubegen.orbital(mol, 'h2o_mo1.cube', mf.mo_coeff[:,0])
In the above example, one can change mf.mo_coeff[:, 0] to other orbitals by changing the orbital index.
Loading a molecule
In the VMD main window, click on File → New Molecule, then click on Browse to select the cube file to load. Finally click on Load.
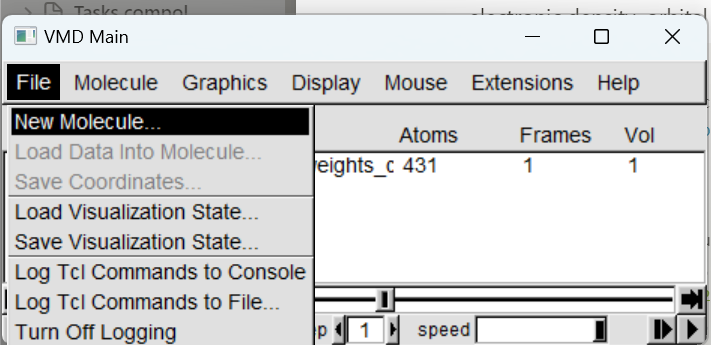
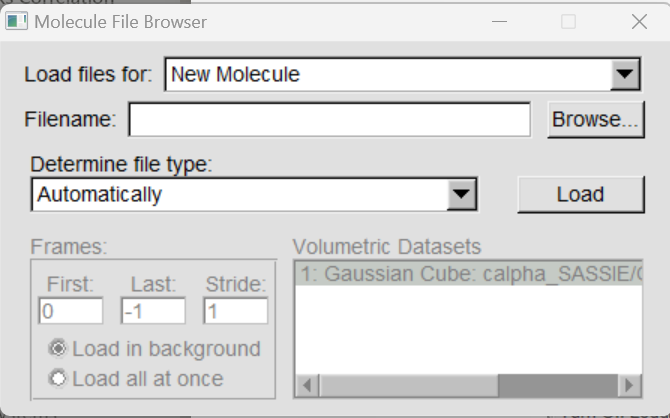
One can keep on loading other .cube files to the current molecule by selecting the molecule, and File → Load Data into Molecule.
Changing the design
The original color scheme can be hard to read, one should change the background and representations for a better view.
Background settings
One can choose the background to be solid color or gradient color in the VMD main: Display → Background.
To change the color, open Graphics → Colors. In the Categories window, select Display, then in the Names window, select Background to change the solid color; BackgroundTop and BackgroundBot to change the gradient color.
Representations of the Molecule
The default of the molecules are dots, and to make the molecules prettier, one needs to change the representations. In VMD Main, open Graphics → Representation.
- Choose the atoms to edit in the Select Atoms window, all means all atoms, type 1 means type 1 atoms, etc.
- Choose the Coloring Method, for molecules, one can choose Element or Mass, and the Drawing Method can be VDW for spheres, CPK for spheres and bonds. One should change the Sphere scale and bond scale to make the best illustration. In the following, we show H2O molecule with VDW (left) and CPK (right) respectively.
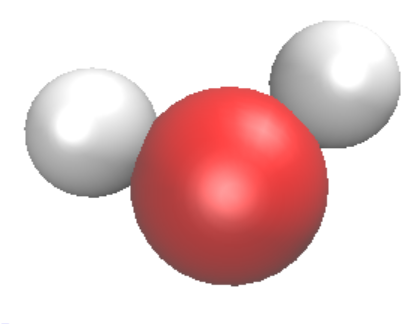
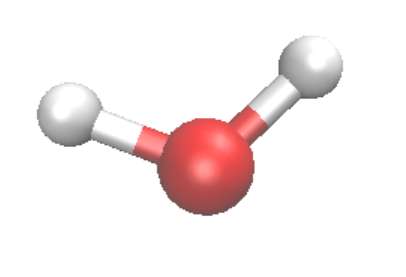
The CPK form can be generated with two layers of representations. One can click on Create Rep to create a new layer and Delete Rep to delete a layer. In order to make the CPK style, one create one layer with VDW, and one layer with Dynamic Bonds.
Plot orbital
The .cube file for orbital information is created by cubegen.orbital . On top of the molecules created above, one can plot the charge density. The following are still in Graphics → Representations.
- Create a new layer, and choose Drawing Method as Isosurface. At the bottom right, choose Draw to be Solid Surface, and Show to be Isosurface.
- Change the Isovalue (one can drag the bar on the right) to change the charge value to show (can be negative or positive). One can also change the Range of the Isovalue. Set Material to be Transparent.
- To assign different colors to positive and negative charge density, create a layer as in Step 2, and choose the Coloring Method to be ColorID. Set Isovalue to be a negative value and choose the number beside the Coloring Method to be, say 0 (Blue). Then create another layer, and set the Isovalue to be positive, and choose another number of color, say 1 (red).
An example of the HOMO of H2O is
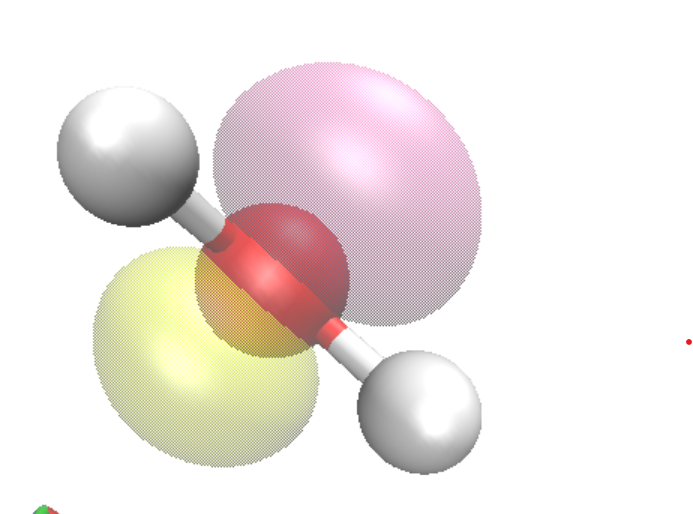
Saving the image
One can save the image to the PostScript (.ps) form. Open File → Render. In the Render the current scene using: window, choose PostScript (vector graphics). One can set the path and name to save.
To convert the .ps file into PDF, in the command line, install ghostview by sudo apt install ghostview . Then use ps2pdf file.ps file.pdf to convert to pdf.